Técnicas de IA para Biologia
10 - Usage of Gene Ontology
André Lamúrias
Usage of Gene Ontology
Inference in the Gene Ontology
Inference in the Gene Ontology
True Path Rule
- Gene Products are annotated to the most specific GO term
- Annotations are (implicitly) propagated to ancestor terms ($\sf is\_a$) as well as via $\sf part\_of$
- True Path Rule: path from a child term through all ancestors back to the root must be biologically accurrate
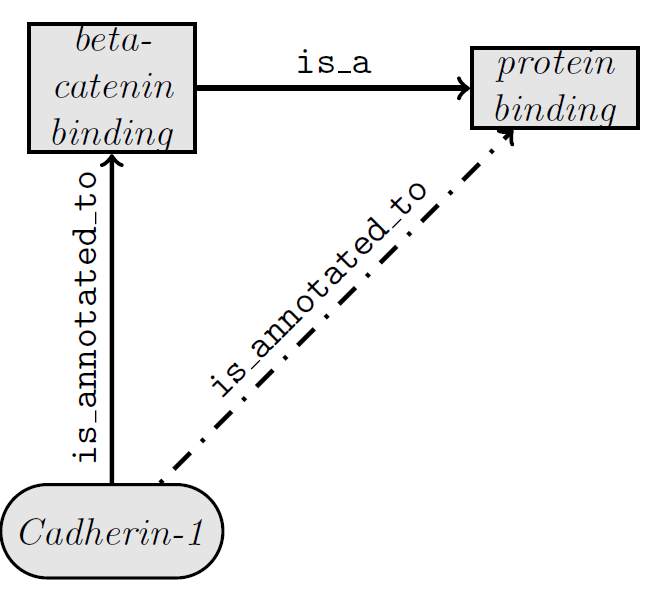
Inference in the Gene Ontology
Problem Example
- For one species (flies): chitin metabolism a child of cuticle synthesis
- But chitin metabolism also part of cell wall organization in yeast
- Yeast gene annotated with chitin biosynthesis implies annotation to cuticle biosynthesis, but yeast does not have cuticles
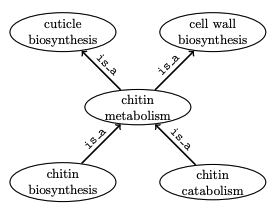
Inference in the Gene Ontology
Problem fix
- Introduction of new GO terms separating the two kinds of processes

Inference in the Gene Ontology
Inferences over GO Edges
- A number of inference rules have been established
- For instance:
$$A \stackrel{is\_a}{\longrightarrow} B \wedge B \stackrel{part\_of}{\longrightarrow} C \Rightarrow A \stackrel{part\_of}{\longrightarrow} C $$
Inference in the Gene Ontology
Example
[Term]
id: GO:0044444
name: cytoplasmic part
is_a: GO:0044424 ! intracellular part
relationship: part_of GO:0005737 ! cytoplasm
[Term]
id: GO:0005737
name: cytoplasm
[Term]
id: GO:0005739
name: mitochondrion
is_a: GO:0044444 ! cytoplasmic part
- We can infer that mitochondrion $\sf part\_of$ cytoplasm holds
Inference in the Gene Ontology
Example
- We can infer that mitochondrion $\sf part\_of$ cytoplasm holds

- This is not the same as saying mitochondrion $\sf is\_a$ cytoplasm
Inference in the Gene Ontology
Other Inference rules
$$A \stackrel{part\_of}{\longrightarrow} B \wedge B \stackrel{is\_a}{\longrightarrow} C \Rightarrow A \stackrel{part\_of}{\longrightarrow} C $$
- Transitivity
$$A \stackrel{is\_a}{\longrightarrow} B \wedge B \stackrel{is\_a}{\longrightarrow} C \Rightarrow A \stackrel{is\_a}{\longrightarrow} C $$
$$A \stackrel{part\_of}{\longrightarrow} B \wedge B \stackrel{part\_of}{\longrightarrow} C \Rightarrow A \stackrel{part\_of}{\longrightarrow} C $$
- Similar rules apply to $\sf has\_part$
- As well as to $\sf regulates$ and its subproperties $\sf positively\_regulates$ and $\sf negatively\_regulates$

Inference in the Gene Ontology
Intra-GO Cross-Product Definitions
[Term]
id: GO:0000152
name: nuclear ubiquitin ligase complex
namespace: cellular_component
def: "A ubiquitin ligase complex found in the nucleus." [GOC:mah]
is_a: GO:0000151 ! ubiquitin ligase complex
is_a: GO:0044428 ! nuclear part
intersection_of: GO:0000151 ! ubiquitin ligase complex
intersection_of: part_of GO:0005634 ! nucleus
- Intersections provide an equivalent definition
$$ GO:0000152 \equiv GO:0000151 \sqcap \exists part\_of.GO:0005634 $$
- Follows the design pattern of a more specific class $X$ (of general class $G$) differentiated by an additional discriminant $D$
- X is a G such that D
- GO:0000152 is a GO:0000151 such that $\exists part\_of.GO:0005634$
Inference in the Gene Ontology
External Cross-Product Definitions
[Term]
id: GO:0001510 ! RNA methylation
intersection_of: GO:0008152 ! metabolic process
intersection_of: OBO_REL:results_in_addition_of CHEBI:32875 !methyl group
intersection_of: OBO_REL:results_in_addition_to CHEBI:33697 !ribonucleic acid
- With external terms from the ChEBI ontology (Chemical Entities of Biological Interest)
Inference in the Gene Ontology
Reasoning with Cross-Product Definitions
[Term]
id: GO:0030223 ! neutrophil differentiation
intersection_of: GO:0030154 ! cell differentiation
intersection_of: OBO_REL:results_in_acquisition_of_features_of\ CL:0000775
! neutrophil
[Term]
id: GO:0030851 ! granulocyte differentiation
intersection_of: GO:0030154 ! cell differentiation
intersection_of: OBO_REL:results_in_acquisition_of_features_of\ CL:0000094
! granulocyte
[Term]
id: CL:0000775
name: neutrophil
is_a: CL:0000094 ! granulocyte
- We can infer that neutrophil differentiation is a subclass of granulocyte differentiation
Usage of Gene Ontology
Overrepresentation Analysis
Overrepresentation Analysis
Overview
- High-throughput technologies in molecular biology
- Allow to measure all genes in the genome experimentally
- DNA microarrays with thousands of probes
- Quantify the amount of corresponding sequences in the sample
- Typical microarray experiments:
- Compare gene expression profiles (their concentrations) under two or more biological conditions
- E.g., comparison between healthy and diseased tissue or different developmental stages
- Several replicate microarray experiments for each biological condition
- Statistical analysis for significant differences for each gene
- Outcome often a list of hundreds/thousands of differentially expressed genes
Overrepresentation Analysis
Idea
- Question: One or more specific GO term annotates more of the differentially expressed genes than one would expect by chance?
- Example
- Say 221 of 6000 yeast genes (3.7%) represented on a microarray are annotated to the GO term sporulation
- If we perform some experiment and observe 100 differentially expressed genes, 3 or 4 should be annotated with sporulation, merely by chance
- Suppose that 35 of 100 are annotated to sporulation
- We conclude that sporulation is overrepresented among differentially expressed genes
Overrepresentation Analysis
Issue
- We may based on such observations develop hypotheses to justify the outcome
- Determine subsequent experiments to test the hypothesis
- Multiple overrepresented GO terms may be indentified, inflating the number of significantly overrepresented terms
- List of 50 to 100 GO terms is not helpful in principle for determining which of the terms is the most characteristic
Overrepresentation Analysis
Solution
- Several algorithms based on hypergeometric distribution and related concepts
- Some on a term for term basis
- Irrelevant terms are omitted upfront (filtered)
- Still often many corrolated terms in results - propagation rule - which one is the most suitable?
- Being annotated to a given GO term, also implies annotation to its ancestors
- Tests for overrepresentation of similar terms are not statistically independent
Overrepresentation Analysis
Solution
- Parent-child algorithms
- Takes the propagation rule into account for determining overrepresentation
- Topology-based algorithms
- Find the most specific overrepresented terms
- Other model-based approaches
- Rather than a term for term analysis, an optimization problem is created that associates a score to a set of GO terms, trying to find an optimal combination of GO terms that together best explain the observed pattern
Usage of Gene Ontology
Semantic Similarity
Semantic Similarity
Overview
- Concepts in ontologies are connected by semantic relations
- Measures of similiarity for terms can be defined based on that
- Used for
- Validating results of gene expression clustering
- Predicting molecular interactions
- Disease gene prioritization
- Clinical diagnostics
Semantic Similarity
Basic Idea - Information Content
- For ontologies of $\sf is\_a$ relations
- Define a propability function $p$ such that $p(C_i)$ is the probability of encountering an instance of class $C_i$
- Recall that $x$ $instance\_of$ $A$ and $A$ $is\_a$ $B$ implies $x$ $instance\_of$ $B$, i.e., $p$ is monotonically increasing as we move to more general concepts
- Unique root $C$ has $p(C)=1$
- Information Content of a term t:
$$ IC(t) = - log\; p(t) $$
- More general concepts provide less information
Semantic Similarity
Probability of a term
- Intrinsic: uses internal structure of the ontology
- e.g. number of descendants / total number of Entities
- Extrinsic: uses frequency on a dataset
- probability that a randomly chosen protein is annotated to $t$, if we choose the protein from the set of all proteins under consideration
- Terms that annotate many genes have low information content
- Terms that annotate few genes have high information content
Semantic Similarity
Example on Information Content
- Annotations on 1000 documents about carnivores
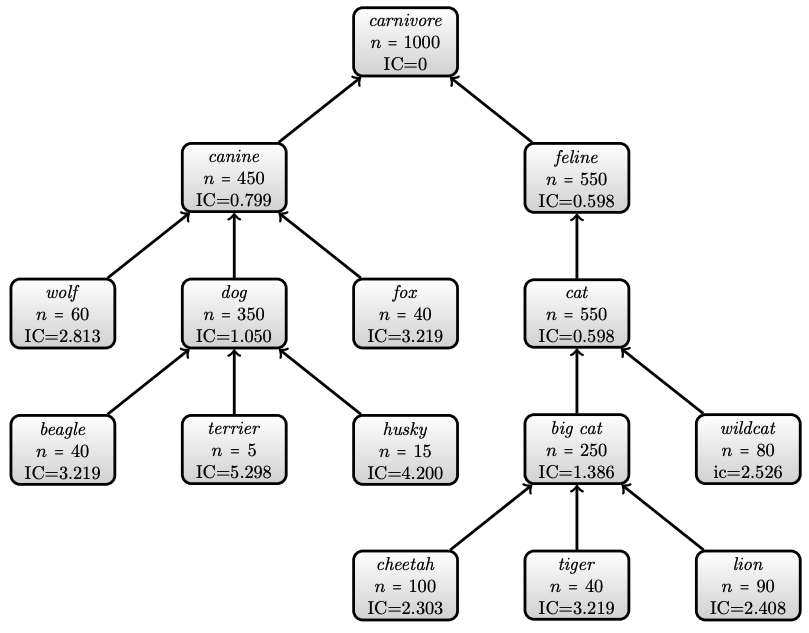
Semantic Similarity
Resnik Semantic Similarity
- The more information two terms share, the more similar they are
- Information shared between two terms is indicated by the information content of their Most Informative Common Ancestor (MICA)
- Similarity between two terms determined as follows:
$$sim(t_1,t_2) = IC(MICA(t_1,t_2)) = max_{t\in Anc(t_1)\cap Anc(t_2)} IC(t)$$
Semantic Similarity
Example on similarity
- Similarity between cheetah and lion

Semantic Similarity
Example on similarity
- Similarity between beagle and wildcat

Semantic Similarity
Improvements
- Variants of similarity measure have been developed
- Take also into account the distance between the terms
- Otherwise wolf and fox are are as similar as beagle and fox
- Can be measured by path length
- But depends on the ontology
- Maximum similarity is reached if the terms are identical
$$ sim_{Lin}(t_1,t_2) = (2 \times max_{t\in Anc(t_1)\cap Anc(t_2)} IC(t))/ (IC(t_1) + IC(t_2))$$
- Further notions have been defined
Semantic Similarity
Applied to GO
- Focus is on similarity between genes annotated by terms - not similarity between terms
- Similarity measures defined based on the similarity of its annotations
- Maximum value of all pairs
- Average
- Also graph-based approaches relying on counting edges
- Set-based measures, e.g., intersection of annotations/union of annotations
Usage of Gene Ontology
Summary
- Inference in the Gene Ontology
- Overrepresentation Analysis
- Semantic Similarity
Further reading:
- Robinson and Bauer, Introduction to Bio-Ontologies, Chapters 8, 10, 12
- Dessimoz and Skunca, The Gene Ontology Handbook
- GO webpage http://geneontology.org/
Exercises
Inference
[Term]
id: GO:0007519
name: skeletal muscle tissue development
is_a: GO:0014706 ! striated muscle tissue development
relationship: part_of GO:0060538 ! skeletal muscle organ\development
[Term]
id: GO:0014706
name: striated muscle tissue development
is_a: GO:0060537 ! muscle tissue development
[Term]
id: GO:0060537
name: muscle tissue development
is_a: GO:0009888 ! tissue development
relationship: part_of GO:0007517 ! muscle organ development
- The CSRPP3 gene is annotated to the GO term skeletal muscle tissue development. What other annotations can we infer for this protein and why?
Exercises
Inference
[Term]
id: GO:0006310
name: DNA recombination
is_a: GO:0006259 ! DNA metabolic process
[Term]
id: GO:0042148
name: strand invasion
is_a: GO:0006259 ! DNA metabolic process
relationship: part_of GO:0006310 ! DNA recombination
[Term]
id: GO:0060542
name: regulation of strand invasion
is_a: GO:0000018 ! regulation of DNA recombination
relationship: regulates GO:0042148 ! strand invasion
[Term]
id: GO:0060543
name: negative regulation of strand invasion
is_a: GO:0045910 ! negative regulation of DNA recombination
is_a: GO:0060542 ! regulation of strand invasion
relationship: negatively_regulates GO:0042148 ! strand invasion
- MPH1 protein is annotated to GO:0060543. What other ...
Exercises
Gene Ontology (Python)
- Download basic version of GO ontology
- http://current.geneontology.org/ontology/go-basic.obo
- Download GO Semantic Similarity file http://labs.rd.ciencias.ulisboa.pt/dishin/go202104.db.gz
- Install:
- pip install goatools ssmpy
Exercises
Gene Ontology (Python) - Basics
from goatools import obo_parser
go_obo = 'go-basic.obo'
# create a dictionary of the GO terms
go = obo_parser.GODag(go_obo)
go_id = 'GO:0048528'
go_term = go[go_id]
print(go_term)
print('GO term name: {}'.format(go_term.name))
print('GO term namespace: {}'.format(go_term.namespace))
for term in go_term.parents:
print(term)
for term in go_term.children:
print(term)
rec = go[go_id]
parents = rec.get_all_parents()
children = rec.get_all_children()
for term in parents.union(children):
print(go[term])
Exercises
Gene Ontology (Python) - common ancestors
- Find the nearest common ancestor of GO:0048527 and GO:0097178
def common_parent_go_ids(terms, go):
# Find candidates from first
rec = go[terms[0]]
candidates = rec.get_all_parents()
candidates.update({terms[0]})
# Find intersection with second to nth term
for term in terms[1:]:
rec = go[term]
parents = rec.get_all_parents()
parents.update({term})
# Find the intersection with the candidates, and update.
candidates.intersection_update(parents)
return candidates
Exercises
Gene Ontology (Python) - common ancestors
def deepest_common_ancestor(terms, go):
# Take the element at maximum depth.
return max(common_parent_go_ids(terms, go), key=lambda t: go[t].depth)
...
go_id_id1_dca = deepest_common_ancestor([go_id, go_id1], go)
print('The nearest common ancestor of\n\t{} ({})\nand\n\t{} ({})\nis\n\t{} ({})'
.format(go_id, go[go_id].name,
go_id1, go[go_id1].name,
go_id_id1_dca, go[go_id_id1_dca].name))
Exercises
Gene Ontology (Python)
- What is the name of the term GO:0097192?
- What is the most specific term that is parent of both GO:0097191 and GO:0038034?
Exercises
Gene Ontology (Python) - semantic similarity
- Calculate the semantic similarity between GO:0048364 (root development) and GO:0048486 (parasympathetic nervous system development) based on the number of branches separating them.
def min_branch_length(go_id1, go_id2, go):
# First get the deepest common ancestor
dca = ...
# Then get the distance from the DCA to each term
dca_depth = go[dca].depth
d1 = go[go_id1].depth - dca_depth
d2 = go[go_id2].depth - dca_depth
# Return the total distance - i.e., to the deepest common ancestor and back.
return ...
Exercises
Gene Ontology (Python) - semantic similarity
def semantic_distance(go_id1, go_id2, go):
return min_branch_length(go_id1, go_id2, go)
def semantic_similarity(go_id1, go_id2, go):
return 1.0 / float(semantic_distance(go_id1, go_id2, go))
...
print('The semantic similarity between terms {} and {} is {}.'.format(
go_id1, go_id2, sim))
Exercises
Gene Ontology (Python) - semantic similarity
- Using DiShIn: https://dishin.readthedocs.io/
- Download and uncompress: http://labs.rd.ciencias.ulisboa.pt/dishin/go202104.db.gz
import ssmpy
ssmpy.semantic_base("go.db")
ssmpy.ssm.intrinsic = True
e1 = ssmpy.get_id(go_id1.replace(":", "_"))
e2 = ssmpy.get_id(go_id2.replace(":", "_"))
print(e1, e2)
print(ssmpy.ssm_resnik(e1,e2))
print(ssmpy.ssm_lin(e1,e2))
Exercises
Gene Ontology (Python) - semantic similarity
- Similarity between proteins
e1 = ssmpy.get_uniprot_annotations("Q12345")
e2 = ssmpy.get_uniprot_annotations("Q12346")
ssmpy.ssm_multiple(ssmpy.ssm_resnik, e1, e2)
ssmpy.ssm_multiple(ssmpy.ssm_lin, e1, e2)







